Speeding KNIME with Voracity
Abstract
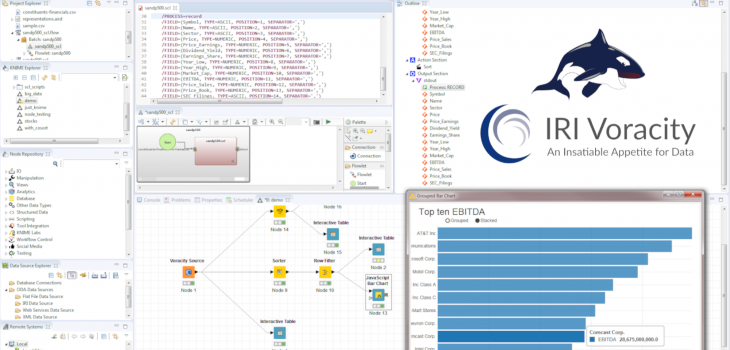
KNIME is a leading open source analytic and visualization tool for data scientists. Wrangling raw data for KNIME projects is usually done via their intermediate file node, database connectors, or other extensions like Spark. To increase functionality and speed while reducing the complexity of data preparation, IRI created a ‘job source’ or ‘data provider’ node to use the CoSort engine. Its SortCL program, also built in the IRI Voracity data management platform and running in Eclipse with KNIME, simultaneously wrangles and feeds integrated, cleansed, masked, or synthesized data in memory into the KNIME workflow. PC benchmarks show that this approach can drive dramatic KNIME performance gains in high volume.
Background
KNIME, short for Konstanz Information Miner, is an open source data mining and analytics platform for turning data from multiple sources into charts, images, data models, and outputs, all in a single workflow designed and run from Eclipse. KNIME has become a very powerful, and popular ecosystem for predictions, machine learning, and other areas of data science.
KNIME projects consist of task-specific nodes, which are all the input, analytic and visualization pieces of the larger or job. KNIME provides many free nodes of its own, and a marketplace of nodes built by the KNIME community. There are nodes for machine and deep learning (AI), predictive analytics, custom code, and nodes for topics like molecular chemistry.
The nodes are strung together in workflows can also run individually or in groups. But as with other BI and analytic platforms, KNIME jobs run slow given large data sources. Its own data sourcing and transformation nodes are inherently slow, and external nodes like Spark are limited in functional and data source scope.
Fortunately, a new node compatible with the IRI Voracity data manipulation and management platform — which is also built on Eclipse but powered by the IRI CoSort data manipulation engine called SortCL — can result in KNIME workflows finishing up to up to 10x faster. CoSort is a proven big data workhorse featuring superior transformation algorithms, optimized I/O and memory use, and task consolidation.1
The “Voracity Job Source” node for KNIME rapidly wrangles raw data using SortCL and pumps its results into a KNIME model in memory. This model can then be used by almost every other node in KNIME. Those nodes can thus work immediately with the filtered, transformed, cleansed, and PII-masked data that they need to succeed. This article covers the relative planning and performance of data wrangling in KNIME without, and then with, the Voracity node.
Benchmarking Environment
For testing the node, I used CSV source files of increasing size2 to compare the relative processing speeds and scalability of the single Voracity Job Source node against the typical KNIME nodes provided to read the same input and perform the same data transformations.
My test files contained four transaction items: department, store number, date, and sales by department. They were chosen to simulate a data set from a department store chain where product sales are tallied by department and by store.
All tests were performed in the IRI Workbench IDE for Voracity with the KNIME Analytics Platform installed in the same Eclipse workspace. I will explain how to load KNIME components into the IRI Workbench along with the Voracity node in another blog post.
The hardware is used for the benchmarks of both approaches was on an Intel hexa-core Windows 10 64-bit desktop with 12GB of RAM. The software versions used are CoSort v10 (in Voracity v2) and KNIME v4.
Prerequisite KNIME Nodes
To perform a group by to find the total sales for each store, I needed to configure, connect, and run five nodes in KNIME to achieve the same result Voracity will using only one node.
The nodes needed for this test:
- CSV Reader: reads the CSV file into KNIME
- Number to String: changes the store numbers to strings
- Column Filter: removes all columns but the store and sales column
- Round Double: sales column from the CSV file was automatically read in as an integer. This data type is too small for the sum in the next node
- GroupBy: Sums the totals of all the sales and groups them by store
- Interactive Table: Shows the results in a table form
These nodes and others can be found in the node repository by either using the search bar or by navigating to, and selected them from, the respective categories in which they appear.
Prerequisite Voracity (CoSort SortCL) Data Wrangling Job
For the Voracity Job Source node to work, there needs to be a preexisting SortCL job script with a stdout (standard output / unnamed pipe) target specification. These jobs are also made in IRI Workbench, through a variety of methods like automatic job creation wizards, ETL-style mapping diagrams, and a syntax aware editor.
In this example, I am sorting and aggregating the test department store transaction data set in different-sized CSV files. This requires a single job script for the SortCL program to parse and run on the command line, or this case, to be launched in the Voracity provider node for KNIME.
My job script is shown in IRI Workbench below, along with its dynamic outline and diagram view:
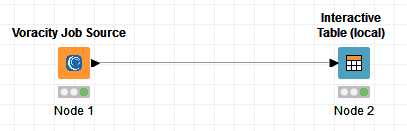
This job is then selected for execution in the one (and only) Voracity Job Source node required:
Note that a SortCL job can not only perform multiple transformations in the same script and I/O pass, it can take multiple sources (files, tables, pipes, URLs, etc.) and produce multiple targets all at the same time. It uses source field names as the symbolic references for mapping the data to the outputs. 3
Remember, the first target you want feeding to KNIME must be designated as stdout (standard output) for the data to flow from SortCL in memory to the KNIME table node. Without that defined pipe, the Voracity node will not work.
Running the Voracity Job Source Node
Using the Voracity Job Source node is similar to using any other node in KNIME. Just go to the IRI Voracity Node category and drag the Voracity Job Source into the workflow:
Open the Node and use the file browser to locate and select the target script (.scl file) that SortCL will run in the Voracity Job Source node for KNIME:
After you select the script, click Apply and run the Voracity node to create and feed the SortCL results directly, through memory, into the target KNIME table. That created table can be transferred to other nodes for analytics, other mining, or visual nodes in KNIME:
Relative Speeds
Using the Global Timer node provided by KNIME, we can compare the speed of KNIME with and without the Voracity node in performing the same work. The listed times are each node’s individual time in milliseconds from the workflow.
The following screenshots show how the normal Global Timer node’s table would appear.
Times for 1 million rows with just KNIME Nodes:
Total time for those nodes:
Times for 1 million rows with Voracity in KNIME:
Total time for Voracity in KNIME:
After various tests on the increasing file sizes, below are my final results. The times listed are the average totals of all nodes involved to create the same table at the end of each workflow.
These times illustrate a major improvement over the functionally equivalent, required KNIME nodes; i.e., what takes seconds for Voracity takes minutes in KNIME.
More specifically, given the same flat-file CSV sources, Voracity wrangled that data up to 9 – 10x faster than KNIME! In fact, KNIME needed more time just to read the CSV file than Voracity needed to complete the entire data transformation job and transfer that data to KNIME.
Bottom Line
By pre-processing and calculating data for KNIME before it has to convert it into its own table, we can greatly increase the speed at which KNIME can perform its total set of data preparation and analytic functions.
In both methods of course, downstream analytics in the workflow proceeds identically, but the major bottleneck associated with them is alleviated when the Voracity node provides the data.
Next Steps
So far, the Voracity node works with KNIME inside native Eclipse builds like, or including, IRI Workbench. Learn how to install it here.
Check out this application of the Voracity node feeding data into KNIME machine learning nodes in the evaluation of breast cancer data. Give us your feedback and suggestions in the reply form below.
- You can read about the foundations of CoSort and the roles of Voracity in big data here.
- Those files were also built in Voracity using SortCL, which is also the parent program for the IRI RowGen random test data synthesis product.
- The task consolidation of its external sortcl engine is one of the reasons Voracity is a superior alternative to legacy ETL tools in its own right.



















1 COMMENT
[…] This article covers installation of the KNIME Analytics Platform as a core feature in the IRI Workbench IDE for Voracity, built on Eclipse, as well as the IRI Voracity Data Provider, or Job Source, node for KNIME described in this article.1 […]